Researchers from the University of Nottingham have developed a way to select which DNA to sequence with the pocket-sized MinION. The new feature speeds up on-the-spot, real-time DNA sequencing.
 Developed by the billion-euro Biotech Oxford Nanopore (UK), the small MinION may not have the accuracy and speed of normal laboratory machines for next generation sequencing (NGS).
Developed by the billion-euro Biotech Oxford Nanopore (UK), the small MinION may not have the accuracy and speed of normal laboratory machines for next generation sequencing (NGS).
But its size and price could bring DNA sequencing to applications that require cheap, fast and on-the-spot results. One example includes the field surveillance of strains of Ebola virus.
The versatility of portable sequencing could now be improved with a new feature. Researchers from the University of Nottingham have developed a way for the MinION to identify if the part of DNA it’s currently ‘reading’ is of interest. If it’s not, it doesn’t bother wasting time on it and starts with the next fragment.
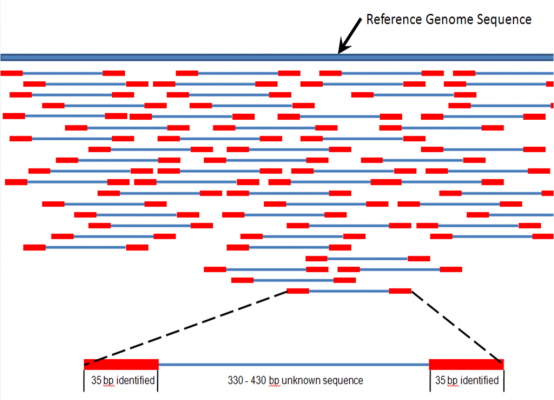
Named ‘Read Until‘, the novel technique was now published in Nature Methods. It’s the first time that direct selection of specific DNA molecules has been shown on any device, and it could greatly reduce the analysis time.
Oxford Nanopore’s devices use molecular pores (a technology now in a patent dispute) that produce electric current fluctuations when DNA fragments go through. These fluctuations were termed ‘squiggles‘, and are identified as a specific base pair (the ‘letters’ of DNA) with software – constructing the fragments’ DNA sequence step by step.
In their research, the team bypassed the individual recognition of DNA base pairs. Instead, it uses signal processing techniques to map the squiggles to reference sequences of DNA. So it’s more like identifying a word or sentence as a whole, rather than reading each letter and then figuring it out.
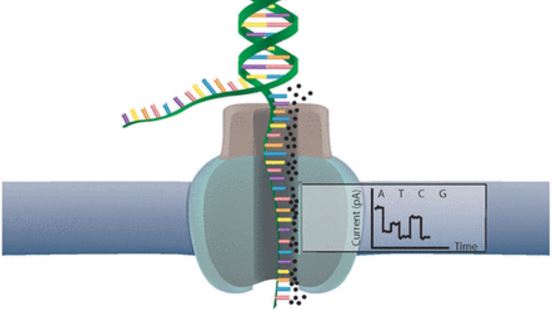
This squiggle matching technique is fast enough to enable real-time decisions, which is where the ‘Read Until’ function comes in. If the part of DNA that was already read by one of the nanopores doesn’t match any sequence of interest, it’s ejected.
Real-time selective DNA sequencing (or more creatively, ‘DNA tasting‘) can be particularly useful to detect pathogens in complex samples (having DNA of the human host, for example) or sequence key DNA fragments. Examples would be to identify strains of infectious diseases or mutations in specific regions of the human genome.
Feature Image Credit: Oxford Nanopore
Figure 1 Credit: Churko et al. (2013) Overview of High Throughput Sequencing Technologies to Elucidate Molecular Pathways in Cardiovascular Diseases. Circulation Research (doi: 10.1161/CIRCRESAHA.113.300939)





