The combination of advanced sequencing technologies and user-friendly data analysis solutions is bringing metagenomics into the reach of more scientists, creating new opportunities to ask and answer exciting research questions.
New ways to look at microbes
Metagenomics is a relatively new field that is radically changing how microbial analyses are performed and making it possible to comprehensively examine the microbial diversity all around us.
Traditionally, microbial genomics relies on culturing organisms and analyzing the resulting nucleic acids by single-genome analysis. But this strategy is highly limited because less than 1% of bacteria can actually be grown in a lab.
As a culture-independent method, metagenomics analyzes the combined DNA sequences of all microorganisms in a community, including the ‘unculturable’ members. This allows researchers to study the composition, function, and dynamics of the complete microbial community, instead of just individual organisms and parts.
Assessing unknown communities

Microbial communities may include bacteria, archaea, fungi, algae, protozoa, and viruses, and they are everywhere: in the soil and oceans, in thermal vents and hot springs, and in drinking water systems. They are all over our bodies, too: on our skin, in our mouths, in our intestines, and reproductive organs.
The compositions and roles of these communities remain mostly a mystery, and understanding them is of great interest to scientists.
Microbial communities affect human health and impact the diagnosis, prevention, and treatment of diseases. They influence plant growth and are important for food crop productivity. They produce much of the oxygen in the air we breathe. Hence, metagenomics finally gives researchers a means to study them.
Some of the first metagenomics research was carried out in the early 2000s, and the availability of more advanced and affordable high-throughput sequencing has fueled the rapid growth in the field. In fact, a literature search shows that in 2010, only 253 research articles mentioned metagenomics, but this number has quickly increased, with over 1000 articles each year since 2017 across all disciplines: life sciences, biomedical sciences, energy, bioremediation, biotech, agriculture, microbial forensics, earth science, and others.
Bridging the knowledge gap with clever computational solutions
One of the biggest challenges in metagenomics is managing the enormous amounts of highly complex sequencing data. It must be filtered, assembled, and then annotated through an iterative process that usually requires an advanced skill set in computational science, statistics, programming, data mining and visualization, database development, and modeling.
Successful data management and analysis are key to harnessing the full power of metagenomics and gaining useful insights. But most microbiologists are not trained in bioinformatics and thus lack the expertise to analyze their data.
Tübingen-based company Computomics is working to bridge this knowledge gap by providing powerful metagenomics data analysis solutions that researchers can easily run on their own without technical hurdles or specialized programming skills.
Meet MEGAN6, the complete metagenomics toolbox
MEGAN6 is their comprehensive, stand-alone metagenomics analysis software. It is currently the only software in the world capable of performing interactive analyses on a regular desktop computer through a graphical user interface.
“Analyzing metagenomics data is an iterative process, and MEGAN6 streamlines the procedure,” explained Christian Dreischer, Scientific Project Manager at Computomics. “You can quickly and easily investigate the differences between individual organisms, microbial communities, or entire microbiomes.”
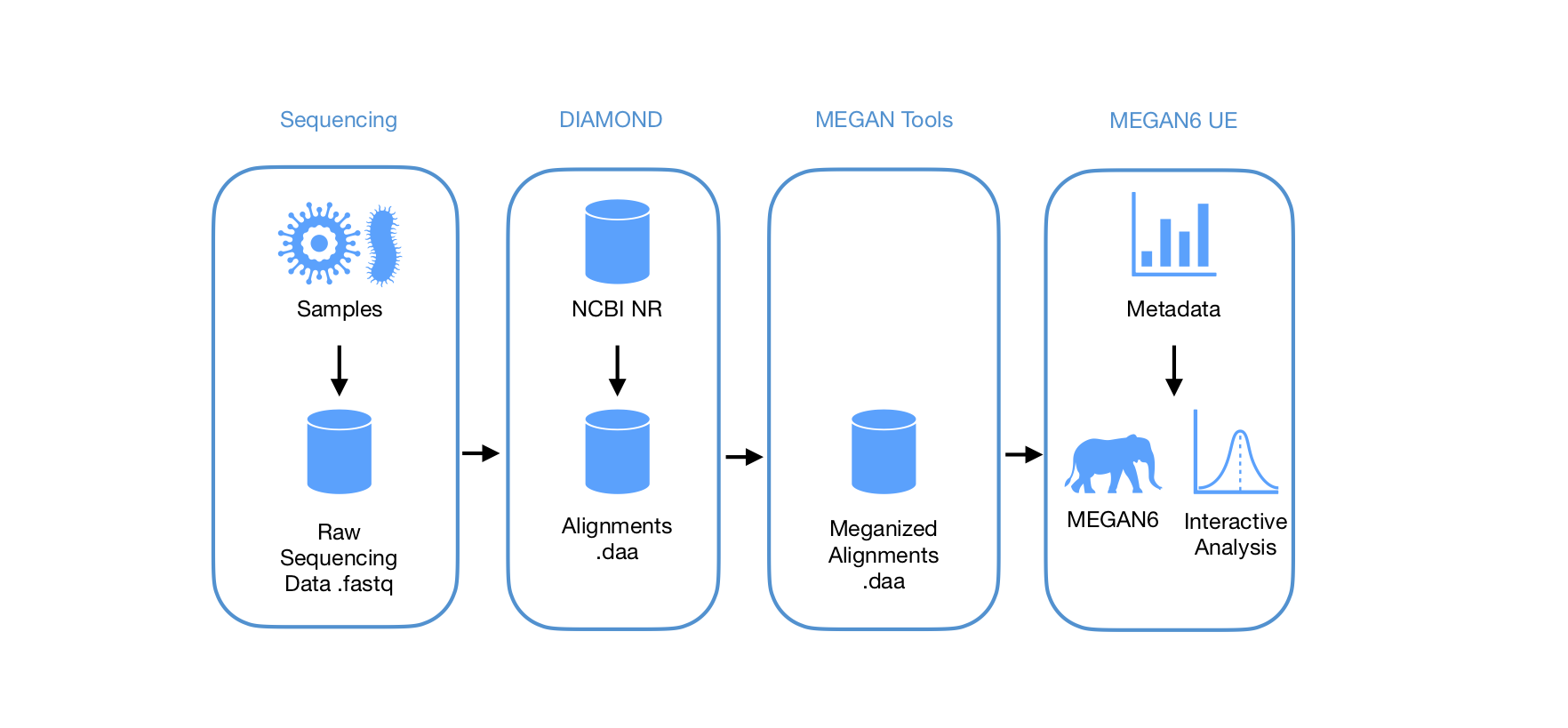
The typical MEGAN6 workflow for whole-genome sequencing samples includes annotation of raw sequence data against the largest public protein reference database (NCBI NR) using the ultra-fast aligner DIAMOND. Alignments can be loaded straight into MEGAN, where interactive analyses are performed. Pre-processing of the DIAMOND and MEGAN tools steps can also be automated using MORPHEUS.
The MEGAN6 Ultimate Edition (UE) includes a license for the KEGG pathway database, which enables researchers to directly explore the functional roles of the communities they are studying and determine not only which biochemical and metabolic pathways are present, but also their differential abundances.
Integrated visualization tools facilitate data interpretation and allow the creation of insightful figures and charts. MEGAN6 UE also allows customization and the development of automated analysis pipelines.
MORPHEUS: a ready-to-use analysis interface
MORPHEUS is Computomics’ web-based metagenomics analysis interface, developed for those who prefer to run useful analyses without dealing with large files or lengthy processes. Scripts for most common analyses and visualizations for taxonomy and function studies are included.
MORPHEUS is simple to use, even for novice metagenomics researchers, and can be customized for more in-depth analyses if needed.
Custom bioinformatics services and data interpretation
Computomics also offers custom services spanning the complete metagenomics workflow so researchers can access the expertise they need to achieve their project goals.
“If you need advice on experimental design, such as the number of samples or the right sequencing depth or technology, we can help. This way you can be sure that the data you collect can actually answer your questions,” says Dreischer. “We can also perform a complete data analysis and deliver a written report to help you get to your insights quickly.”
Asking and answering the deeper questions
Metagenomics analysis software MEGAN is cited in over 4000 research articles, ranging from assessing the impact of antibiotic treatment on the gut bacteria resistome to profiling antibiotic resistance genes in drinking water reservoirs, and even determining the composition of the Iceman Ötzi’s gut microbes and final meal.
These comprehensive but still user-friendly tools allow researchers to draw innovative insights from complex datasets, enabling them to answer deeper questions: How diverse are microbial communities in the environment? Are they random, or do they have similar patterns? Do they interact with each other? What are they doing and why? Someday soon, we will know.
Would you like to find out more about Computomics’ data analysis tools? Listen to this podcast about MEGAN6 or visit their website to get more information!
Header image via Shutterstock.com and article images via Computomics
This article was originally published in October 2020 and has since been updated.