The research team led by Emannuelle Charpentier, co-discoverer of the CRISPR/Cas9 system, has found that a new CRISPR system can also cleave RNA. Amazingly simple!
 The discovery of the CRISPR/Cas9 system revolutionised Biotechnology, unlocking a kind of ‘Golden age’ for gene editing – with a wide-reaching impact from therapeutics to SynBio.
The discovery of the CRISPR/Cas9 system revolutionised Biotechnology, unlocking a kind of ‘Golden age’ for gene editing – with a wide-reaching impact from therapeutics to SynBio.
So, it’s no wonder that the scientists that invented CRISPR are on the road to win all the scientific prizes – though who did ‘invent’ it is still a controversial issue.
However, outside of Science’s red carpet bubble, research on CRISPR continues. One of the biggest breakthroughs was that Zhang’s team (at the Broad Institute) described a new protein with similar properties to Cas9.
This ‘second’ cutting protein is Cpf1, which has some key advantages over Cas9. Namely, it is smaller, only requires one RNA strand (instead of two) and targets different sites, opening up a wider range of applications.
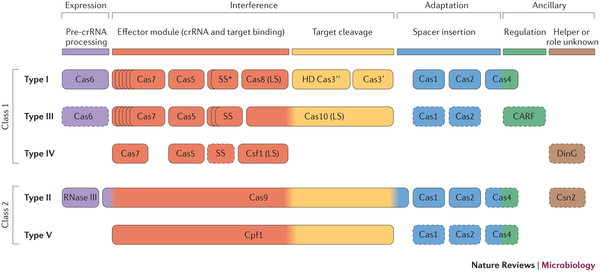
Now, the most minimal CRISPR system yet has been described in a new paper published in Nature.
This CRISPR/Cpf1 system was found in Francisella novicida, and boasts a remarkable feature. The data suggests that the Cpf1 protein has a dual function, with separate domains to cleave not only DNA (the original characteristic that enabled the first example of gene editing), but also RNA.
This means we are now able to cleave sections of pre-crRNA to make it into mature crRNA. Since RNA is typically described as the ‘Template’ for DNA replications, Cpf1 can make its own template for gene editing – as well as cutting DNA at specific points.
This means it doesn’t require assisting molecules like tracrRNA or RNase III – essentially acting as a multi-purpose DNA editing tool. All in one!
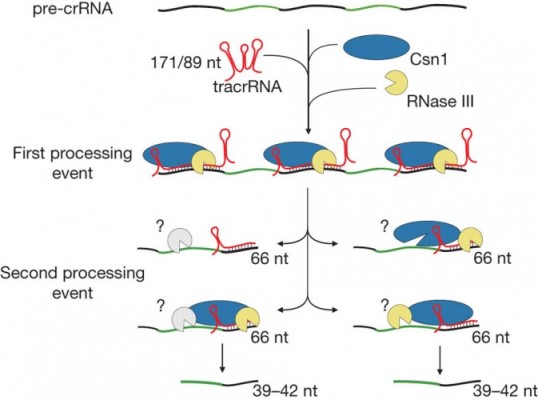
As Charpentier explains,
CRISPR-Cpf1 is a plug-and-play system with no additional component needed.”
This mechanism was discovered by the new team led by Emannuelle Charpentier, which includes researchers from the Max Planck Institute for Infection Biology (Berlin, Germany), Umeå University (Sweden) and the Helmholtz Centre for Infection Research (Braunschweig, Germany).
This is a big deal for the Gene Editing world – as simplifying the process makes it easier to use CRISPR in therapies (as it’s more compact of a system) and could have really exciting applications in the future.





