In response to new treatments, cancer came back stronger and more resistant than ever. Medicine’s counterattack will be more effective with a clear target.
How do you solve a problem like drug resistance in cancer research? There’s no clear-cut answer yet, not least because the disease can vary so widely. But there has been some exciting research working around drug resistance e.g. lung cancer therapies.
As an example, EGFR is overexpressed in a number of cancers, including squamous cell lung cancer. The first generation of drugs, tyrosine kinase inhibitors (TKI’s), worked by checking its up-regulation. However, tumors quickly developed resistance to these treatments, presenting a new challenge to medical researchers.
It was discovered that a mutation, T790M, is present in over 50% of TKI-resistant tumors: consequently, it became the target of second- and third-generation lung cancer therapies. But this leaves almost half of drug-resistant lung cancers without explanation or a target, so there is a nearly 50% chance that the second line of treatment will be ineffective.
A biopsy of the tumor is usually the primary means of analysis, but these are invasive and often only single-use: there is not enough genetic material to do a second test as confirmation of the result. Alternative methods to test for drug resistance mutations are therefore being investigated, and the most promising thus far is cell-free DNA (cfDNA) analysis.
In this method, blood/plasma is collected from the patients’ venous blood and prepared for the extraction of cancer cell DNA. However, cfDNA is fragmented and present only in tiny amounts; and DNA from other cells is also circulating, which make the DNA belonging to the tumor even more difficult to detect. These factors lead to the necessity of a really sensitive system to extract the DNA and then analyze it with digital PCR and/or next-generation sequencing.
PCR is well-established, and there are solid options for NGS workflows for various suppliers but reliable extraction of cfDNA is still a key bottleneck within this workflow.
Sample Volume
Since it’s difficult to obtain substantial quantities of cfDNA from plasma, it would be best to select an extractor that can handle the largest sample sizes possible. The highest volume currently available is 10 mL from PerkinElmer.
Efficiency
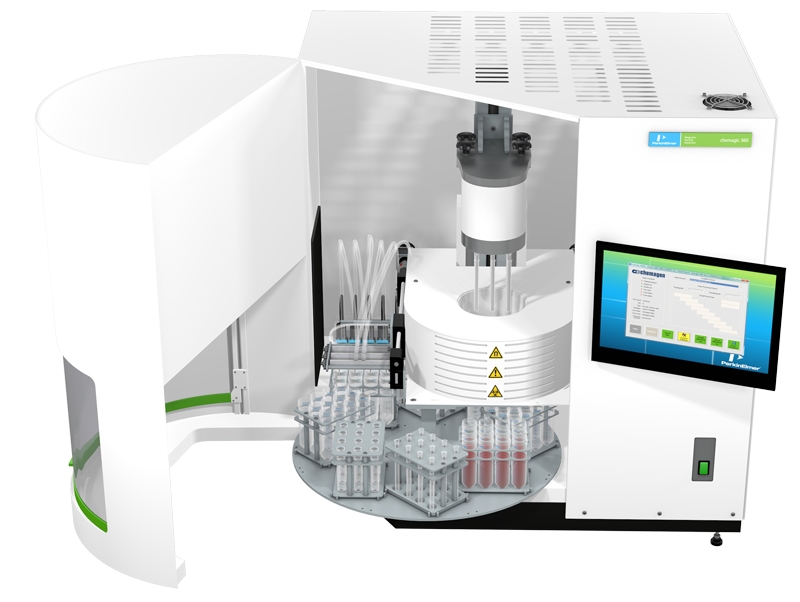
How much bang do you get for your tissue buck? Given that your goal is to extract as much cfDNA as possible, high speed and throughput can be valuable features. With the chemagic 360 research instrument, PerkinElmer allows the extraction of up to 5 ng cfDNA/ml plasma in fast processing times of 24 samples from 2 – 5 mL sample volume within 120 minutes and 12 samples from 8-10 mL sample volume within 90 minutes.
Downstream Suitability
As test related to cfDNA usually require quality-demanding downstream applications like real time/digital PCR, methylation analysis, array technologies, NGS workflows, reliable nucleic acid extraction as the initial step is unalterable. Furthermore, maximum flexibility in sample throughput and automation-grade like the use of liquid handling robotics are of great benefit.

Knowing your target will greatly bolster your effort to undermine a cancer’s drug resistance. A good nucleic acid extraction instrument with mutation identification from cfDNA can save you time and anxiety over whether or not your results are reliable. If you want to know more about chemagic 360 or think this reliable high volume, high throughput nucleic acid extractor sounds like a good investment for you, drop PerkinElmer a line!
For Research Use Only. Not for use in Diagnostic Procedures. US: For Laboratory Use Only. Not Intended For Use in Diagnostic Procedures.
Images: Srirat, Romaset, anyaivanova / shutterstock.com





