By Oxford BioDynamics
Cancer care is in urgent need of effective tests to determine the best approach for personalized medicine. Practical, new solutions exploiting the power of 3D genomics are emerging.
Cancer is a vastly complex, multidimensional disease marked by the uncontrolled proliferation of cells. Many intertwined layers of biological systems give rise to the clinical hallmarks of cancer. Deciphering this tangled network is a challenge, and it obscures our understanding of early cancer detection, cancer progression, and discovering new preventative and therapeutic interventions. Improved biomarkers can help to understand disease drivers and better classify a patient by their probable disease risk, prognosis, or likely response to treatment.
The prevailing “multi-omics” approach generates ever larger, expensive, high-dimensional datasets of many types, including genomic, epigenomic, transcriptomic, proteomic, and metabolic profiles. It relies on the belief that subsequent machine learning data analyses will capture meaningful insights from this data, despite there often being considerable, naturally high, biological variability and noise.
Aided by rapid developments in high-throughput technologies, such as next-generation sequencing, this approach produces an unparalleled volume of data. The integration and interpretation of these huge datasets to deduce useful information is complicated and increasingly requires large-scale collaborations. Indiscriminately adding more data can be counter-productive and get in the way of uncovering actionable insights. If the data has low clinical relevance, this puts a growing burden on statisticians and computational algorithms to cut through the escalating noise.
Despite great effort and resources thrown at developing validated biomarkers for better-classifying patients for precision medicine, cancer biology has defied any effective, reliable solutions.
3D genomics: The great simplifier
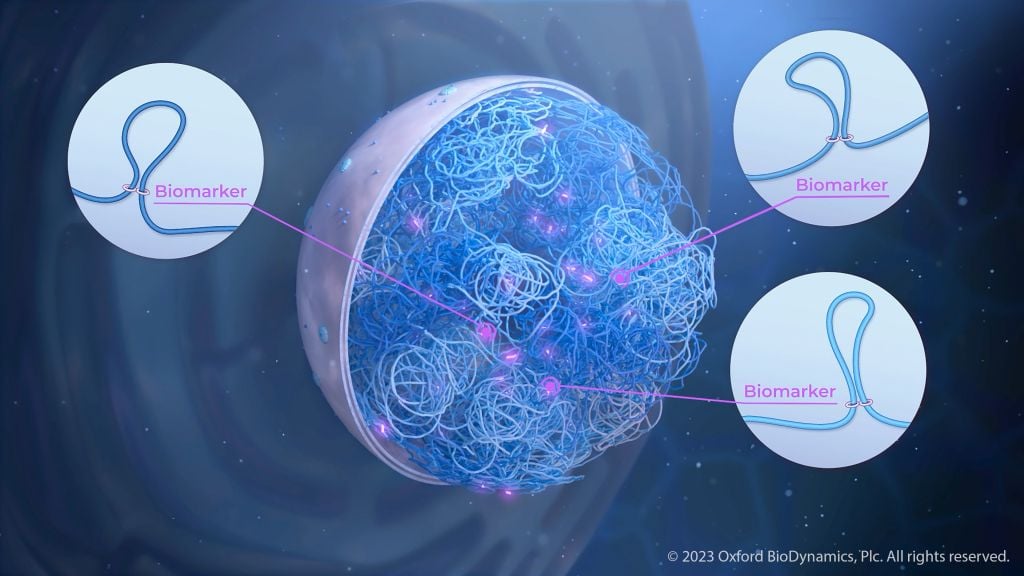
Almost every cell in our body contains two meters of DNA, which is intricately folded to fit in the cell nucleus. This three-dimensional organization of DNA, your 3D genome, appears to be as important as the genetic code itself in controlling cellular destiny and offers a wealth of untapped information relevant to health and clinical outcomes.
Looking at 3D genomics offers fundamental advantages. 3D genomic organization represents an integration of many multiomic signals1. Genetic, epigenetic, transcriptomic, proteomic, and metabolomic alterations can all be reflected in specific 3D genomic changes. The 3D genomic shape, in turn, acts as a powerful regulatory gatekeeper controlling how gene activity is modulated. DNA is folded into loops bringing distant parts of the linear genome into close proximity, thereby influencing each other. These loops are generally stable but can act as switches that undergo changes in response to influences from genetics, environmental cues, metabolism, and cell-to-cell communication2.
Importantly, common 3D genomic patterns can often be identified that are universally and uniquely shared across certain diseases. This is because they are highly prevalent binary events, with a high signal-to-noise ratio. This approach is highly informative for patient classification.
3D genomics bridges the link between multiomic complexity and the clinical phenotype. Over the last 10 years, mounting evidence has shown that 3D genomic profiles can be effective biomarkers3. With the right technology and methodology, it can boil complex biological layers of regulation down to a straightforward series of markers providing robust stratification of clinical outcomes for challenging diseases.
Looking at the system, not just the cancer
Cancer is a systemic disease4. Immuno-oncology has greatly reinforced this notion. The immune system and microenvironment surrounding a tumor play a large part in determining whether cancer spreads, stabilizes, or responds to therapy. Biomarker approaches that look exclusively at a tumor biopsy miss this critical information.
It is well documented that when a set of genetic loci acquire 3D genomic changes, this is not only seen in circulating tumor cells, but also in circulating white blood cells, even during the earliest stages of cancer5. These alterations in immune cells represent systemic changes linked to the cancer and can be used to identify tell-tale information about a distant tumor. In other words, a systemic barcode6 made up of a set of binary 3D genomic biomarkers (a chromosome conformation signature, CCS) can serve as a convenient liquid biopsy biomarker.
The art of knowing where to look and what to measure
For years, analyzing 3D genomic interactions has involved a family of methods called chromosome conformation capture (“3C”, or its more common universal derivation, Hi-C). These, again, generate vast quantities of data and rely on deep sequencing to capture the presence of meaningful markers. However, the space of possibilities of 3D genomic interactions is vast. Without a way of filtering, these methods inescapably pick up many non-specific interactions that are transient and clinically meaningless.
This introduces a very high level of random noise, dwarfing the signal from key regulatory loops. Low sensitivity and reproducibility result in high data mining costs and a lower complexity dataset, which have limited this approach to research applications.
The story so far
Overcoming these limitations and “reducing to practice” an end-to-end platform for discovery, development, and commercial clinical operation of 3D genomic assays is what drove the founders of Oxford BioDynamics (OBD), an Anglo-American biotech, to develop its EpiSwitch platform. The EpiSwitch Explorer Array, a commercial whole-genome microarray built in collaboration with Agilent, can simultaneously interrogate ~1 million potential 3D genomic interactions.
The high-throughput array is encoded with probes that only select for highly reproducible 3D genomic markers, thereby generating rich, clinically meaningful data. Once significant marker leads are identified, these are translated into a MIQE-compliant qPCR format, undergo feature reduction to a minimal signature, are validated on independent clinical cohorts, and can then be tech-transferred for independent validation and operation at a CLIA-lab using standard equipment.
Using established EpiSwitch technology and methodology, OBD has now developed its own portfolio of tests, with the aim of enabling physicians to easily test and classify patients using only a blood test. The first cancer test to use 3D genomics was launched in 2022 – the EpiSwitch CiRT (Checkpoint inhibitor Response Test).
It is a smart blood test for cancer patients that provides guidance on navigating the toughest challenges of immunotherapy, such as treatment planning, pseudo-progression, and adverse events7. The first-of-its-kind test predicts, with 85% accuracy, an individual’s therapeutic response to immune checkpoint inhibitors (ICIs), a family of widely used immunotherapies that give some patients a real boost to their cancer recovery and survival.
While they can offer unprecedented extension of life, only around one in four patients see an overall anti-cancer benefit, and many are kept on the drug despite a lack of positive outcome, significant expense, and up to a 40% risk of immune-related side effects, which can be severe8.
By exploiting systemic 3D genomics, which incorporates signals from the host immune landscape, CiRT has demonstrated best-in-class performance across more than 14 broad oncological indications9. The test is commercially available as a US CLIA-lab service and is steadily being adopted by practicing oncologists, surgeons, and interventional radiologists.
OBD believes that the adoption of 3D genomic testing will enable precision medicines, such as immuno-oncology treatments, to be more effective, safer, and accessible by allowing them to be used more efficiently on the patients who are likely to respond to them best.
References
1: Tordini, F., et al. (2016). The genome conformation as an integrator of multi-omic data: The example of damage spreading in cancer. Frontiers in Genetics, 7. https://doi.org/10.3389/fgene.2016.00194
2: Alshaker, H., et al. (2022). Monocytes acquire prostate cancer specific chromatin conformations upon indirect co-culture with prostate cancer cells. Front. Oncol., 12. https://doi.org/10.3389/fonc.2022.990842
3: Crutchley, J. L., et al. (2010). Chromatin conformation signatures: Ideal human disease biomarkers? In Biomarkers in Medicine (Vol. 4, Issue 4). https://doi.org/10.2217/bmm.10.68
4: Coussens, L. M., & Werb, Z. (2002). Inflammation and cancer. In Nature (Vol. 420, Issue 6917). https://doi.org/10.1038/nature01322
5: Jakub, J. W., et al. (2015). A pilot study of chromosomal aberrations and epigenetic changes in peripheral blood samples to identify patients with melanoma. Melanoma Research, 25(5). https://doi.org/10.1097/CMR.0000000000000182
6: Bastonini, E., et al. (2014). Chromatin barcodes as biomarkers for melanoma. Pigment Cell and Melanoma Research, 27(5). https://doi.org/10.1111/pcmr.12258
7: Oxford BioDynamics Plc. (2022). EpiSwitch CiRT. https://www.mycirt.com
8: Zhao, B., et al. (2020). Efficacy of PD-1/PD-L1 blockade monotherapy in clinical trials. Therapeutic Advances in Medical Oncology, 12. https://doi.org/10.1177/1758835920937612
9: Hunter, E., et al. (2021). Development and validation of blood-based predictive biomarkers for response to PD-(L)-1 checkpoint inhibitors: evidence of a universal systemic core of 3D immunogenetic profiling across multiple oncological indications. MedRxiv, 2021.12.21.21268094.