Newsletter Signup - Under Article / In Page
"*" indicates required fields
Proteins, the key players within cells. They carry out enzymatic functions, are involved in signaling cascades, contribute to the structure of cells, and much, much more. Their abundance and intricate functions make the study of the proteome highly complex. However, once decoded, proteins can be highly useful for the discovery and development of novel therapies, personalized medicines, and biomarker validation and application.
“The proteome is much more than simply an abundance of proteins,” says Christoph Schaab, Vice President of Proteomics at Evotec. “Proteins and their functions are determined by their compartment-specific localization within cells, splicing events, different post-translational modifications, and interactions with other proteins. All of this adds to the complexity of the proteome, but also provides valuable information for researchers.”
Throughout the drug discovery and development process, proteomics can support researchers in a variety of processes. These include analyzing novel targets, understanding disease mechanisms, structure-based drug design, and discovering the mode of action of lead compounds. Moreover, recent advances in mass spectrometry, sample preparation, and bioinformatics have greatly increased the throughput and sensitivity of protein studies.
Learn about biomarker discovery and how they can help us develop personalized medicines
Sensitivity is a key challenge in proteomics

Nevertheless, the complexity of the proteome results in a number of challenges for researchers. “First, the quantification of low-abundant proteins is highly intricate, making sensitivity a key issue,” Schaab explains. “And advancing technologies are continuously delivering greater amounts of data, which means that data interpretation and data integration are also becoming more and more important and difficult.”
Current technologies can cover between 80% and 90% of expressed proteins in experiments, says Schaab. However, proteins low in abundance might play an important role in cell signaling or could be receptors or transporters, for example. Hence, it is essential to analyze less abundant proteins as well.
“Proteomics is really an art,” says Schaab. “You cannot simply buy any instrument or kit and perform proteomics. You can try, but you will not reach the same coverage as the best academic groups out there. Sensitivity depends a lot on the fine-tuning of instruments, on the details in sample preparation, on powerful software tools, on correctly analyzing the raw data. This is something only a few labs in the world can perform at a high level.”
Investing in technologies and expertise is essential to succeed in proteomics
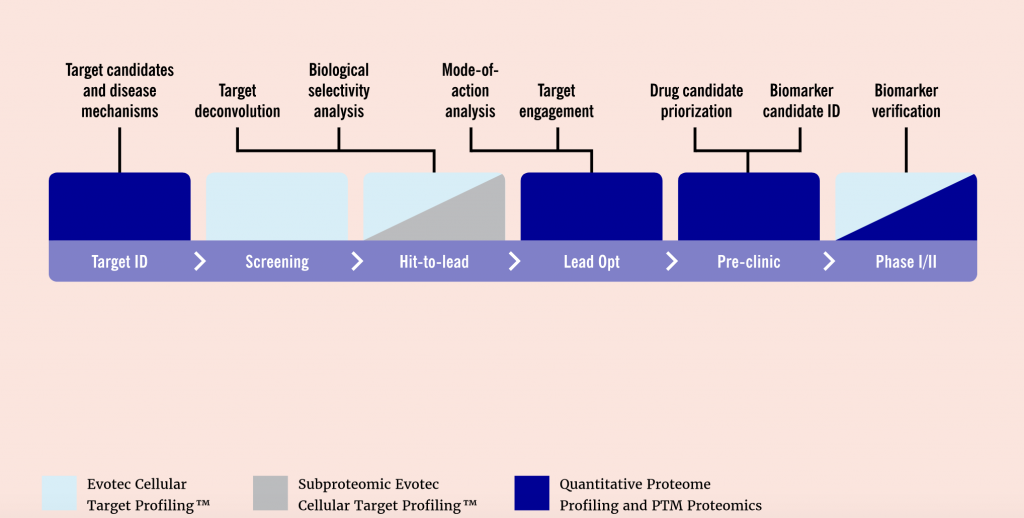
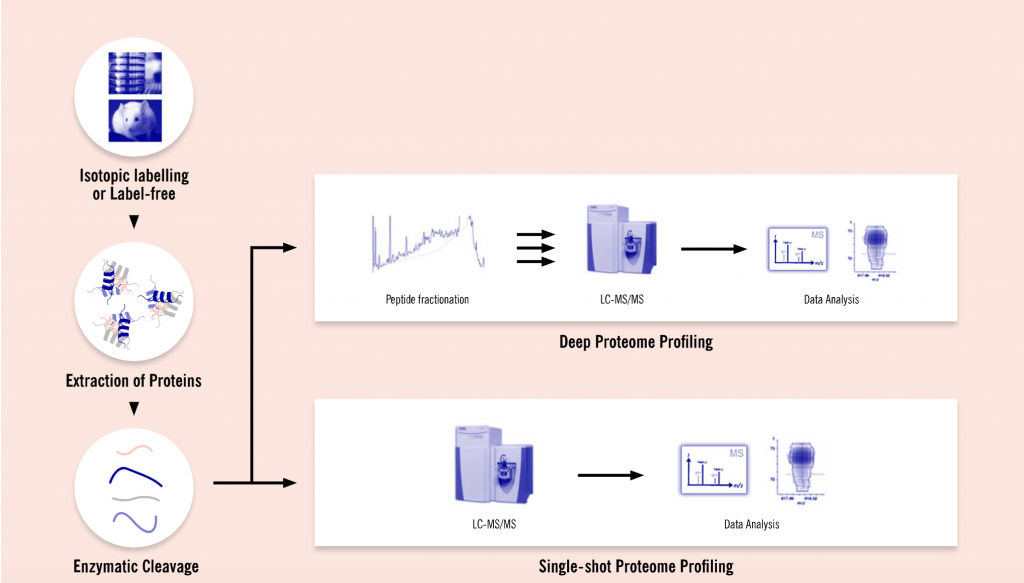
In order to increase the sensitivity of their proteome studies, Evotec has invested in new state-of-the-art instruments and in developing new methods for sample preparation. The company can now support their customers with a tailor-made process starting from experimental design, sample generation and preparation to liquid chromatography-mass spectrometry (LC-MS) measurements, and data and bioinformatic analyses. Upon completion of the project, Evotec’s clients are provided with a detailed report of all experimental procedures, results, and conclusions.
The company’s technologies not only enable the analysis of the proteome but also that of the phosphoproteome and the acetylome, for instance. Moreover, all teams, including the proteomics and metabolomics teams, are integrated within Evotec. This enables close collaboration between teams as well as follow-up studies and the integration of proteomics within the complete drug discovery pipeline.
Like in other research areas, proteomics is also generating increasingly complex data. “Not only is the data becoming more complex, it is also getting more interesting to combine and integrate the data with other omics technologies, like transcriptomics or metabolomics,” says Schaab. “This means we have to develop new algorithms for data analysis and interpretation. In the end, we don’t want our proteomics experiments to generate tables with thousands of data points, we want to know what is actually happening in the cell, what pathways are involved and what is the effect on the cell. At Evotec, we are developing algorithms that can answer these questions precisely.”
Strong ties to important players in the proteomics field and beyond
The proteomics group at Evotec was founded in 2006 as a spin-off from the Max-Planck-Institute of Biochemistry in Martinsried, Germany, by Professor Axel Ullrich and his colleagues. Now Emeritus Scientific Member of the institute, Ullrich focused mainly on kinase inhibitors and immuno-oncology at the time.
Since then, the proteomics group at Evotec has worked with a variety of collaborators from the industry and academia. Their connection to the Max-Planck-Institute is still strong. Here, they work closely together with Professor Jürgen Cox, head of the research group Computational Systems Biochemistry and with the Department of Proteomics and Signal Transduction lead by Professor Matthias Mann.
The capabilities of the group were further expanded by the acquisition of a former Sanofi research site that focuses on proteomics and metabolomics in Toulouse, France. “Evotec has a really broad spectrum of people with very different expertise,” says Schaab. “Many people have worked in the proteomics field for ten or more years. Others are experts in biochemistry, chemical proteomics, and bioinformatics. This has allowed us to expand our technology portfolio to other applications. For instance, we’ve studied numerous posttranslational modifications, invested in biomarker discovery studies, and recently also in MHC peptidomics.”
Using proteomics for biomarker discovery
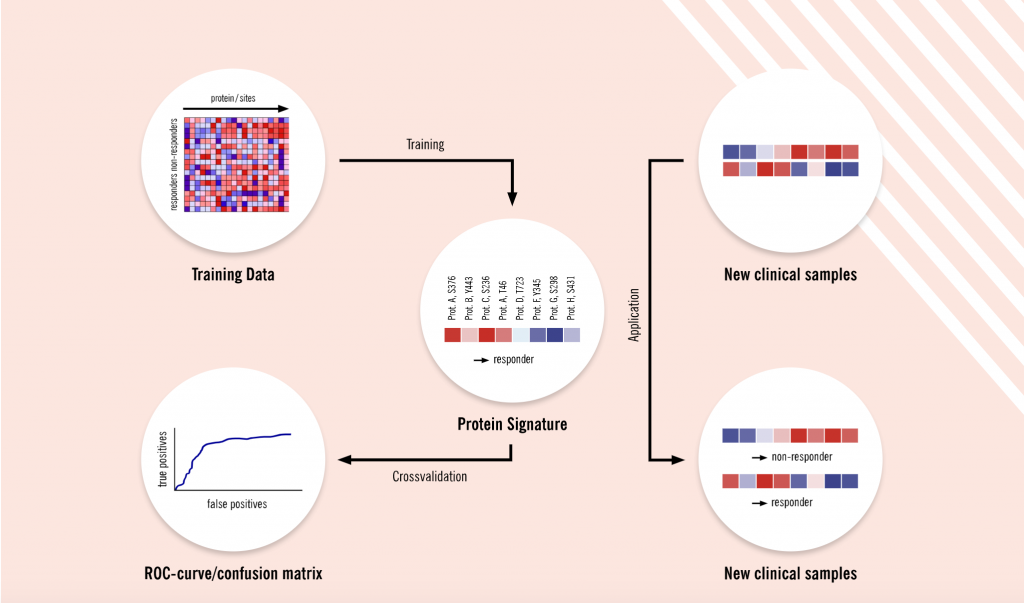
Proteomics allows researchers to make a data-driven, unbiased and hypothesis-free comparison of proteomes, for instance, between healthy and diseased states, treated and untreated or responsive and non-responsive groups.
“At Evotec, we have been looking at the global proteome, not just targeting a few proteins,” says Schaab. “We are studying a combination of multiple features called protein signatures, which are more predictive than single markers. We have already demonstrated that our proteomics platform, combined with advanced machine learning methods, can be used to discover these predictive protein- and phospho-signatures. Initially, we focused on signatures that can predict responses to cancer treatment. In parallel, we’ve also performed larger studies with our partners from pharma and academia to identify pharmacodynamic markers in blood and urine.”
Enabled by advances in technology, robotic automation of sample preparation, data analytics, and bioinformatics, Evotec can now apply proteomics on a global scale in unbiased studies of thousands of samples in an industrialized setting.
Chemical proteomics has a wide field of applications
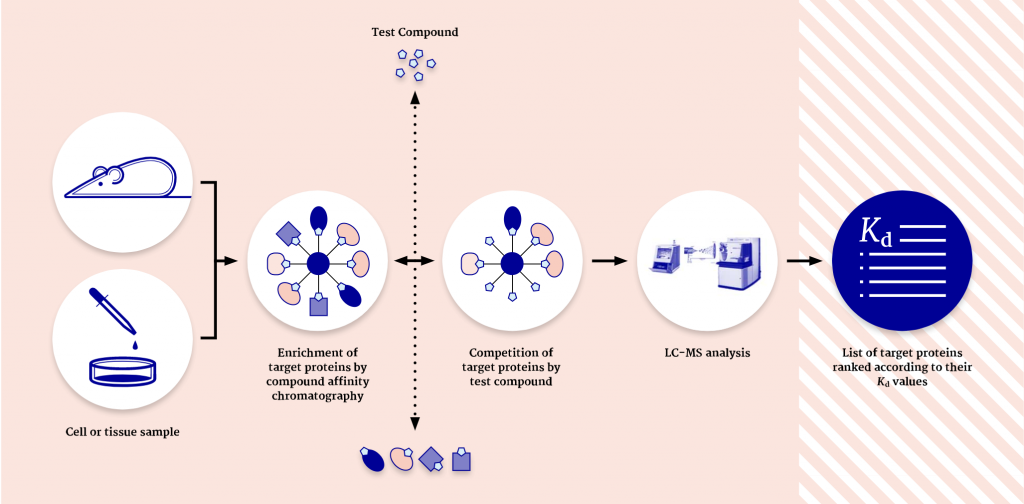
One of the main applications of chemical proteomics is target deconvolution, a process in which molecular targets of active hits are identified. However, the method is also used for target profiling of lead compounds, especially when there is a spectrum of interesting targets or off-targets that have to be identified.
Moreover, later in the pipeline, chemical proteomics can be used for drug repurposing when it is important to understand the different targets that bind to a small molecule of interest.
“At Evotec, we are using three main approaches,” Schaab explains. “Cellular target profiling, drug photoaffinity labeling, and KinAffinity®. With all three methods, we analyze the native, endogenously expressed, and posttranslationally modified proteins in the presence of their cellular cofactors and native complex partners. This is not an in vitro assay but is looked at in a cellular context. This method leads to a higher sensitivity than other methods and also allows us to identify targets with low abundance and low affinity.”
The future of proteomics
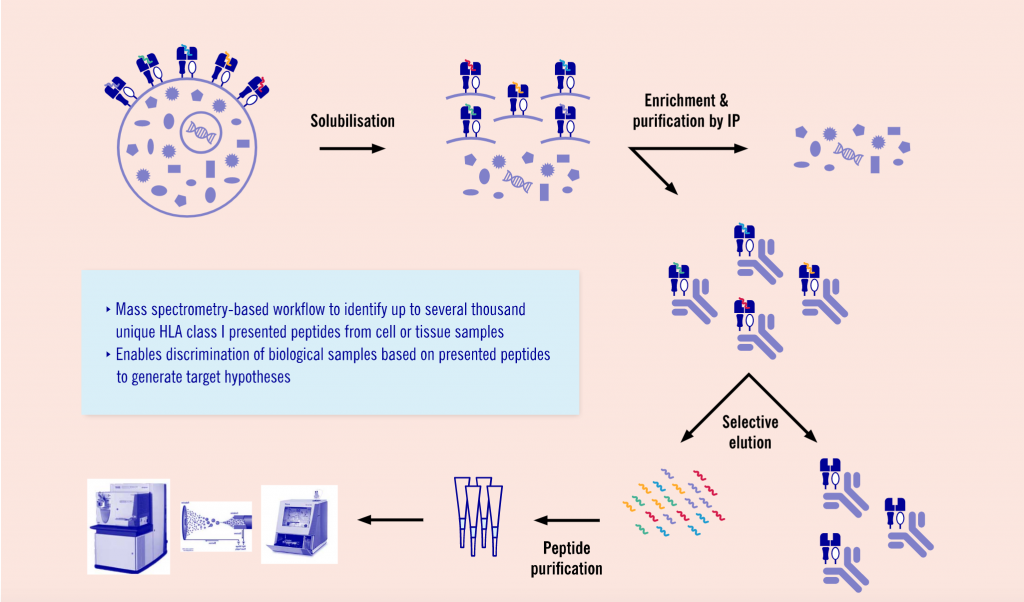
Another promising and upcoming field Evotec is working in is major histocompatibility complex (MHC) peptidomics. This area of proteomics, allows researchers to understand the antigens presented by MHC molecules ‒ an important aspect of immuno-oncology and infectious diseases.
“MHC peptidomics is currently one of the hottest applications of proteomics,” says Schaab. “Very recently, our colleagues at the Max-Planck-Institute published a workflow for analyzing these MHC and presented peptides. Based on this, we established and further optimized the workflow to analyze these peptides. We can combine the results with deep sequencing data, which enables the identification of neo antigens ‒ so peptides that are only presented by tumors and not by healthy cells and tissues.”
In the future, Schaab sees further advances in technical instruments and improvements in their sensitivity, speed, and cost, which will result in larger studies with shorter time frames.
“Progress in sensitivity, especially, will lead us to the point where we will be capable of analyzing single cells using proteomics,” he says. “First steps into the proteome analysis of single cells have already been made by Budnik et al. but the cells were still quite large and the proteome coverage relatively low. In the next few years, sensitivity will further increase and with better instruments, we will arrive at cell proteomics at some point. This will be very exciting.”
Are you interested in learning more about proteomics and its use in biomarker discovery or MHC peptidomics? Download more information here or get in touch with Evotec’s proteomics group and benefit from their expertise at info@evotec.com!
Images via Evotec & header image via Shutterstock.com
Author: Larissa Warneck, Science Journalist, Labiotech.eu