Newsletter Signup - Under Article / In Page
"*" indicates required fields
A research team from the LKS Faculty of Medicine, the University of Hong Kong (HKUMed) has developed a new way to break through the current limited throughput in optimizing precise genome editors at scale, engineer hundreds of base editor variants in parallel instead of current one-by-one testing, and informing users of the most suitable ones for therapeutic genome editing.
The finding has been published in Cell Systems and a patent application has been filed based on the work.
Background
Base editing is a newer CRISPR-based genome editing technology and is a safer tool for tackling genetic diseases with single-base mutations (such as sickle cell disease, familial hypercholesterolemia, etc.) in DNA by correcting them back to their normal form. However, existing base editing can result in different outcomes depending on the type and version of the base editor used, the sequence composition of the target DNA, and the position of the DNA base(s) to-be-converted.
Picking a suboptimal base editor for application can generate incorrect edits and extra mutations around the target DNA base, which could cause undesired effects. Currently, one-by-one testing has to be done to characterize the editing performance of the available base editors to optimize their use on each therapeutic locus. In addition, many therapeutic loci do not have an existing optimized base editor for precise editing yet. Despite worldwide efforts, creating a new base editor can take months or years using conventional methods.
Research findings
HKUMed’s research team successfully created a platform coupling a base editor reporter system with CombiSEAL, an existing technology, to quickly engineer hundreds (or more) of base editor variants in parallel, with the combinations of varying enzymatic deaminase domain and CRISPR/ Cas9-based DNA-recognition domain.
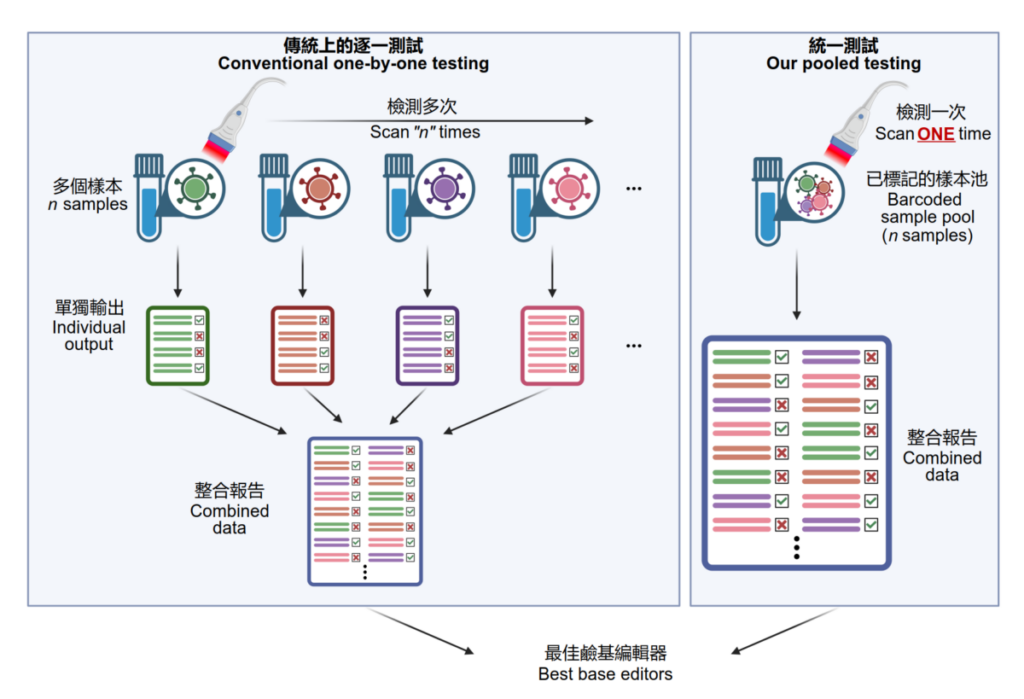
The compatibility and performance of these variants were yet to be characterized and compared in a head-to-head manner. The team applied the platform to quantitatively read out each variant’s editing efficiency, purity, sequence motif preference, and bias in generating single and multiple base conversions in human cells, which helps select the most suitable ones for therapeutic target by generating a particular type of base conversion with maximal efficiency and minimal undesired edits.
The team extended the use of the platform to further enhance the efficiency of the current base editor system. The team members performed a screen focusing on engineering the stem-loop-2 region of the sgRNA (a single guide RNA) scaffold used in the base editor system, and successfully identified two novel sgRNA scaffold variants, SV48 and SV240, that outperform the wild-type scaffold to achieve greater (up to 2.2-fold higher) base editing efficiency.
The team also demonstrated that the platform not only can be used for base editor characterization and screening, but also is compatible with other precise genome editor systems such as prime editors. This could expand the scope of the work in search for other suitable editors to correct genetic mutations at therapeutic targets where a base editor is not applicable.
Research significance
“It is like an accelerated check-out process in stores. Since all product items (i.e. base editor variants) are tagged with a barcode, when it comes to the check-out counter barcode scanner, we need to only put all items in bulk into the basket at the check-out counter. The scanner can automatically identify all items and complete the payment (i.e. base editing performance analysis in our case). There is no need to individually test each base editor one-by-one,” said Alan Wong Siu-lun, associate professor of the School of Biomedical Sciences, HKUMed.
The research was led by Wong Siu-lun. John Fong Hoi-chun, PhD student, was first author, with assistance from Chu Hoi-yee and Zhou Peng, postdoctoral fellows, School of Biomedical Sciences, HKUMed.
Partnering 2030: The Biotech Perspective 2023







