Newsletter Signup - Under Article / In Page
"*" indicates required fields
Contrary to popular belief, Francis Mojica was the first person to discover the CRISPR mechanism and give it its name. He told me how the discovery came about, discussed the wide potential of the gene editing tool and shared his favorite CRISPR application.
You’ve likely heard of CRISPR/Cas9, a gene editing tool that has taken the world by storm. Although it’s far from being the first tool to edit DNA, CRISPR technology makes gene editing much easier and faster than before. In the last few years, its use has become ubiquitous in labs around the world.
CRISPR/Cas9 is used to speed up drug discovery and the creation of animal models to study all sorts of diseases. But the application everyone is avidly expecting and throwing money at is using CRISPR to cure human diseases. The first clinical trials have already started, and their results will reveal whether CRISPR can fulfill these big expectations.
Jennifer Doudna, Emmanuelle Charpentier and Feng Zhang, co-inventors of CRISPR/Cas9, are today the faces everyone associates with the gene editing tool. But the one who discovered the mechanism at the core of the tool and who gave CRISPR its name is Francis Mojica, a Professor at the University of Alicante, Spain.
I was really excited to speak with the person that, years ago, planted the seed that eventually sprouted the whole movement around CRISPR today. I used the opportunity to ask him about everything from the story of how he discovered the basis of such a revolutionary tool to his favorite CRISPR application and what he thinks about its real potential as a therapy.
Let’s start from the beginning. When did you encounter CRISPR for the first time?
It was in 1992, while I was working on my thesis. I worked with microorganisms of the archaea group that live in environments with high salinity. They are extreme halophiles, or ‘salt lovers’. We investigated the mechanisms of adaptation to changes in salt concentration.
We started to sequence their DNA. Back then it was the start of sequencing and you could only read about 200 base pairs if the whole experiment went really well. We eventually found these regularly spaced repeats, which at first we called tandem repeats, or TREPs. We looked at the literature to see if they also appeared in other microorganisms. Nowadays, that is something very easy to look up, but back then there was no PubMed, so we had to search manually, which was tremendously complicated.
In the end, we found that in the year 1987, a Japanese group had encountered repeats of more or less the same size as those we had seen in halophiles and were regularly spaced, which was a unique characteristic.
That got us interested in researching what their function could be. The size of the genome of prokaryotes is very limited, they cannot allow themselves any luxuries. Meaning that if they dedicated such large regions of DNA to maintain these repeats, they must have an important function. And if they were in both bacteria and archaea, the origin of these repeats had to be ancestral.

At first these repeats had several different names. Why did you name them CRISPR in the end?
The first name was TREPs, from tandem repeats, which was a suggestion from my thesis supervisor. But I didn’t think it was the right name. They weren’t really in tandem, which implies they are contiguous, with the same sequence repeated without anything in between. In 2000 we confirmed that equivalent repeats were present in many other organisms, very far away from each other in terms of evolution, and I called them short regularly spaced repeats, or SRSRs.
At the same time, there was a research group — the only back then working with these repeats — that was using these regions as an identification method for Mycobacterium tuberculosis, equivalent to how forensic medicine identifies to whom a human biological sample belongs. They had been using the term DR, for direct repeats.
They contacted me in 2001 to tell me they had found some genes right next to these repeats in different organisms and wanted to agree on a common term. I proposed several alternatives and we went for CRISPR, which stands for clustered regularly interspaced short palindromic repeats. Shortly after, the group published a paper that described the cas genes, an abbreviation for CRISPR-associated.
We now know that CRISPR and these cas genes are part of a primitive immune system of bacteria to cut and kill DNA from parasites. How did you realize this was their function?
After my postdoc, I came back to Alicante and started a research group dedicated exclusively to studying the function of these repeats. We were studying CRISPR as genetic markers of E. coli bacteria. In one of the samples, we found a DNA sequence that was identical to that of a virus. And not any virus, but one that normally infects E. coli, which suggested that this sequence was taken up by an ancestor of this bacteria when it was infected by this virus.
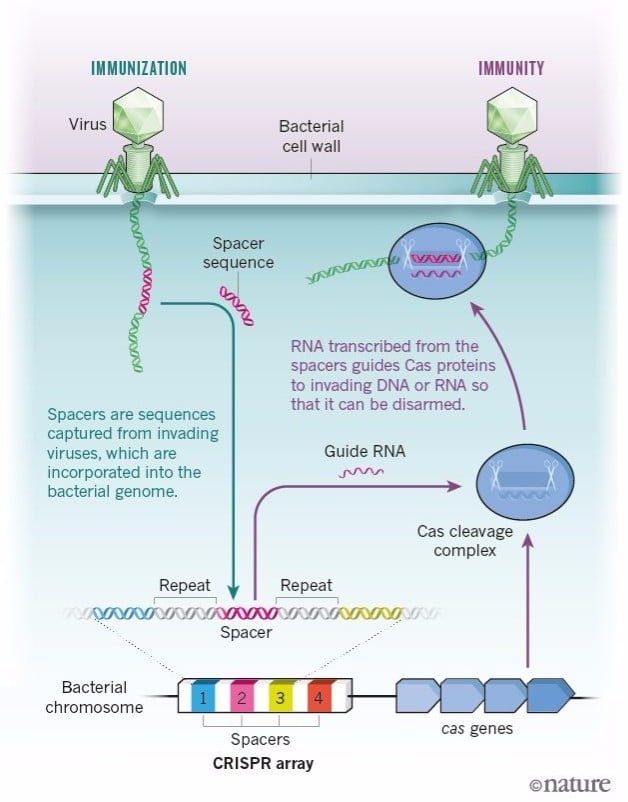
It took us about 6 years to find that out. It was only in 1995 that the whole genome of a bacteria was sequenced. And at the end of the 90s there were maybe 20 bacterial genomes sequenced, whereas now we have thousands.
But in 2003, DNA sequence databases were starting to grow exponentially. Sequences similar or identical to that one, not only from E. coli but from many other, very different organisms, started to pop up. And in all cases, they coincided with the sequences of parasites that infected that precise organism.
Looking up in the literature, we saw that the presence of these DNA sequences seemed to protect the cell from infection by a virus with that same sequence in its genome. We saw there were no cases described of viruses being able to infect cells that had the sequence. Meaning it immunized the bacteria or archaea that carried it.
That was very evident for us, so we tried to publish in Nature, but they said that it was not that interesting. That if we had any proof of what the mechanism could be they might think about it. We evidently didn’t agree.
Well, Nature probably regrets it now…
They were actually not the only ones that didn’t want to publish it. We sent it to four more journals and, in the end, we published in one that, let’s say, was not in the top 10.
Still, there seemed to be a race to be the first to publish this discovery.
Well, four journals said no one after the other, and the one that ended up publishing it took 6 months to get back to us. Back then, submissions were not via the internet, but by mail. After 6 months, we did the changes requested in 2 or 3 days and they took another 3 months until they accepted it. Then they took another 3 months to publish it.
Imagine you have something you know it’s big in your hands and there’s the possibility that another article that takes the originality of your work is published. I remember sending mail every month to the editor, saying ‘please tell me if you’re going to publish or not so we can submit to another journal’. I was in absolute despair.
In fact, a month after publishing, there was another article that said that equivalent DNA sequences of Yersinia pestis, the bacteria responsible for plague, proceeded from viruses. I recently contacted the authors of this article and they said they had also been trying to publish for over a year.
But while the other article was only in one species, we analyzed over 60 strains of around 50 species of different taxonomy groups. It was an exhaustive and complete analysis.
Let’s fast-forward to 2012, when Charpentier and Doudna published their paper on using CRISPR as a gene editing tool. Were you expecting something like this?
No, not at all. The article we initially sent to Nature said our discovery was going to have a big impact on biotechnology, biology and clinical sciences. However, we were thinking about bacteria used to produce pharmaceuticals or for food fermentation because the discovery allowed to select or program these bacteria to be immune to viruses.
But when we saw in 2012 that they had identified the components needed to program a pair of scissors that could be programmed for genome editing, that perplexed me. I am a microbiologist of prokaryotes. When you cut the DNA of a bacteria, you kill it. But I didn’t know that in eukaryotes, when you cut DNA you activate a repair mechanism that opens the possibility to rewrite DNA.
Jennifer and Emmanuelle did it in vitro and it worked wonderfully. A few months later there were two articles — from Feng Zhang and George Church — published in the same journal, back to back, that reported the use of these tools for the first time to edit the genome of mice and humans.
What’s your point of view on the current patent dispute between them?
It is understandable to a certain extent. But it is a fight between two institutions, mainly. I know the researchers personally and I’m convinced they didn’t have the intention to initiate a battle, at least at this level.

Did you ever consider filing a patent yourself?
It crossed my mind but I didn’t. It was never a priority for me. And it wouldn’t have been possible anyway, we didn’t have the experimental proof to make the necessary claims. Though I could have patented the name CRISPR, in which case I could now be a billionaire. But it doesn’t keep me awake at night, someone would have probably changed the name to not interfere.
All the popularity and money CRISPR has attracted is mostly due to the promise of using it as a therapy. Do you think CRISPR can really cure genetic diseases?
What’s sure is that it is greatly advancing the knowledge of the causes of disease in model animals. That is an indisputable reality, and has been done for the last few years.
CRISPR has been used in animals, especially in mice, to cure retinitis pigmentosa, muscular dystrophy, Huntington’s disease, hemophilia, and to eliminate HIV. But in humans, no results yet. We will know in a few months whether it can be used as a therapy.
What is your favorite CRISPR application?
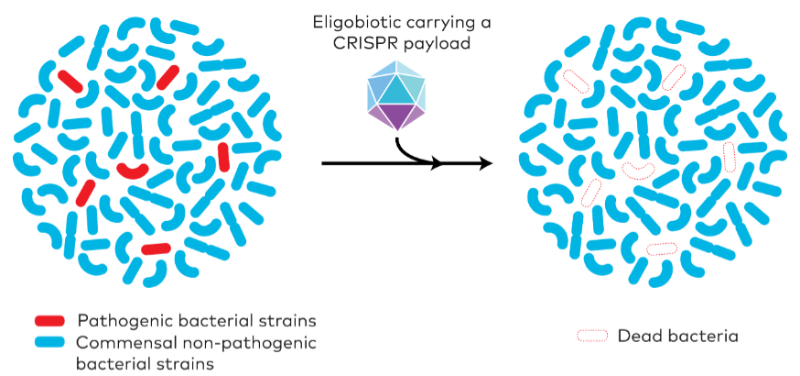
Well, everyone is worried about health, but as a microbiologist I am biased. For me, one of the most interesting applications is as antimicrobials to kill bacteria. These tools can be programmed to specifically target the DNA of bacteria that are pathogens. This has advantages for the gut microbiota – if you take an antibiotic you kill all the other beneficial bacteria in the gut.
You’ve been praised as a possible candidate for the Nobel prize for the discovery of CRISPR for the last few years, but it still hasn’t arrived. Do you think CRISPR will eventually get the Nobel prize?
I am convinced it will get it. When? I don’t know. Nobel prizes are not given the next day. In some cases, it takes even decades until the work is recognized. However, in the case of the PCR, it was given after a few years.
It is possible they are waiting for CRISPR to demonstrate all the potential that is expected, but that would be unfair. What CRISPR has already achieved is much more than what other tools that have received the Nobel have achieved. The prize has gone to tools used to cut and copy DNA in the test tube. CRISPR can be used to edit genomes, change expression levels, visualize DNA, kill bacteria, develop diagnostics, and many more applications, even to store a movie within DNA.
What are your plans in case you do win the Nobel?
Actually, I wish they would have given the prize to CRISPR already, even to others, so the huge pressure I am under would end. I now spend my life taking hundreds of requests that arrive continuously. It’s already been several years and instead of fading it’s getting bigger.
There is no proof I’m even nominated because it’s not public, but if I get it I will disappear from the planet. I need to rest and relax, and I need time to get back to what motivates me and return to the lab.
Pictures of Francis Mojica via Roberto Ruiz /Taller de Imagen de la Universidad de Alicante; Images via NIK Spencer/Nature; The Conversation; Eligo Bioscience. This article was originally published on November 2017.






